Elucidation of antimicrobial resistance mechanisms in pathogenic bacteria

Nosocomial, or hospital-acquired, infections are an important problem worldwide that affects both developed and developing countries. Nosocomial infections are a major cause of mortality in patients admitted to the hospital and have become a public health problem. Multidrug resistant bacteria are commonly the causative agents of nosocomial infections. These infections are difficult to treat and often require the use of new generation antibiotics, which are highly expensive. The research that is being conducted in the Laboratory of Biotechnology is performed using the bacterial species: Pseudomonas aeruginosa and Stenotrophomonas maltophilia. These pathogens, which are often multidrug resistant, are among the most common causal agents of nosocomial infections in Thailand.
Research is being conducted to characterize the novel mechanisms underlying multidrug resistance in these bacteria using genetic engineering techniques to isolate genes that are involved in antibiotic resistance. The results of the research can aid in the identification of new drug targets. The Laboratory of Biotechnology has discovered the novel multidrug efflux transporters, MfsA and MfsQ, from S. maltophilia. MfsA contributes to the transport of several groups of antibiotics, especially those belonging to the fluoroquinolones, out of the bacterial cells, thereby rendering S. maltophilia resistant to multiple antibiotics (Fig. B). MfsQ also functions as an efflux transporter and is involved in bacterial resistance to quaternary ammonium compounds, which are widely used as disinfectants. P. aeruginosa also possesses multiple drug efflux pumps such as Mfs1 and Mfs2. High expression of Mfs1 and Mfs2 is associated with increased resistance to antibiotics belonging to the aminoglycoside, quinolone and cephalosporin families. They function by triggering ArmZ-MexZ interaction. This interaction leads to increased production of MexXY, an RND-type antibiotic efflux pump system and may serve as a target for the development of antimicrobial therapies.
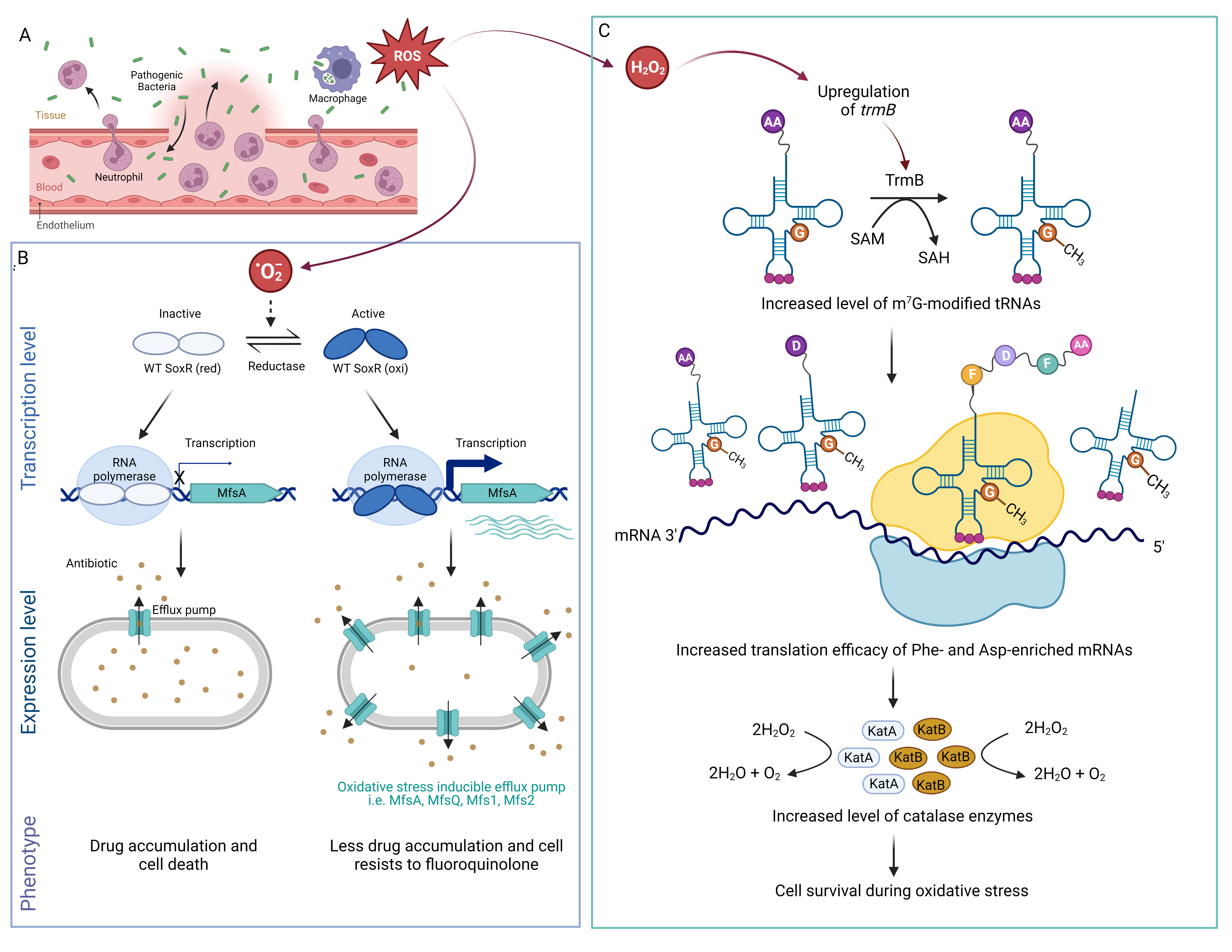
Figure. A schematic of the host response to bacterial infection showing the production of ROS by immune cells (A); the bacterial response to oxidative stress during host-microbe interaction in which superoxide anion (·O2–) activates SoxR leading to increased mfsA expression levels resulting in a fluoroquinolone resistance phenotype (B). Hydrogen peroxide (H2O2) up-regulates TrmB production, thus increasing the level of m7G-modified tRNA, which enhances translation of Phe and Asp enriched H2O2 detoxifying enzymes resulting in cell survival during oxidative stress (C).





